Getting a full picture of how spatial information is processed in the brain has always been challenging because of the need to study multiple brain regions at the same time. Now, a collaborative team led by researchers at Physics for Medicine Paris and Iconeus has shown that functional ultrasound (fUS) imaging can provide the necessary brain-wide information in rats, enabling correlations with speed of movement to be uncovered, and information flows to be mapped.
Over the last 50 years, we have made substantial progress in understanding how navigation abilities are encoded in the brain. We now appreciate the role of various types of neurons in processing information about an animal’s location within an environment, and in updating that internal ‘map’ as the animal moves about.
However, some aspects of the mechanism remain poorly understood – especially the relationships between individual neurons and larger-scale phenomena such as hippocampal theta rhythms, and how information ‘flows’ across the whole spatial navigation system.
A new article published in the journal Cell Reports, and authored by a team including staff from Iconeus and Physics for Medicine Paris, now fills in a gap in this understanding. By mapping how blood volume (a proxy for neuronal activity) changes across the brain as an animal navigates through its environment, they’ve been able to uncover correlations with the animal’s speed and position, as well as shed light on the transfer of information across the brain.
Brain-wide studies with fUS
One of the challenges in investigating the spatial navigation network in the brain is the need to acquire information across multiple areas simultaneously, particularly in the entorhinal cortex, parahippocampal and hippocampal region where spatial cues are known to be processed. This is difficult for traditional electrophysiology, which because it uses electrodes, is inherently limited in its spatial resolution.
The team realized that fUS, with its high spatial resolution (100 μm), high temporal resolution (80 ms), and ability to acquire brain-wide scans of awake, freely-moving rodents, would be an excellent tool to investigate the changing patterns of cerebral blood volume (CBV) – and hence neuron activation – as animals moved within their environment.
To achieve this, the team used an Iconeus One fUS system with a compact IcoPrime-Lite probe in a mobile setup, with the rats able to freely explore a square arena with sides of length 1 m.
Robust correlations of CBV and animal speed
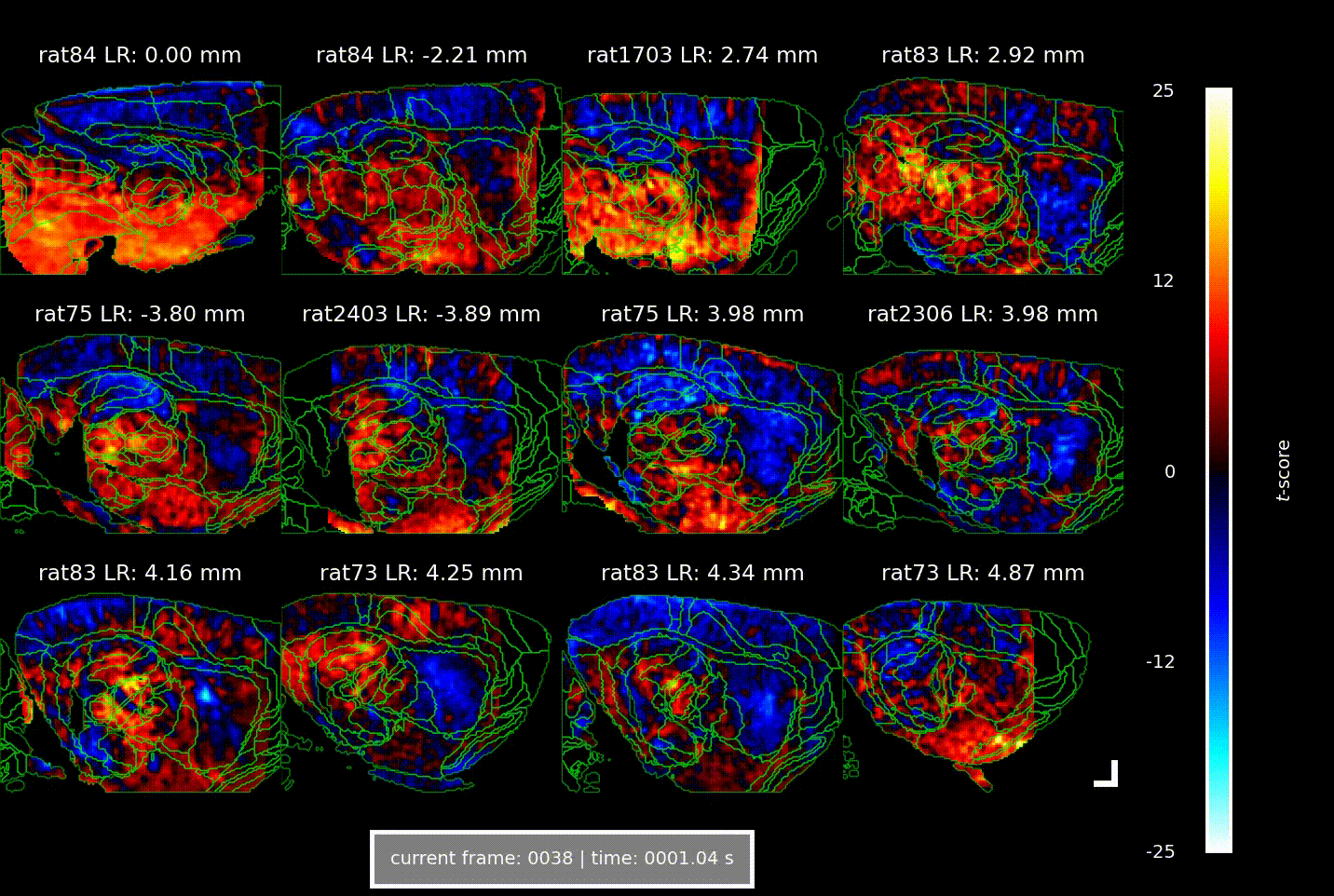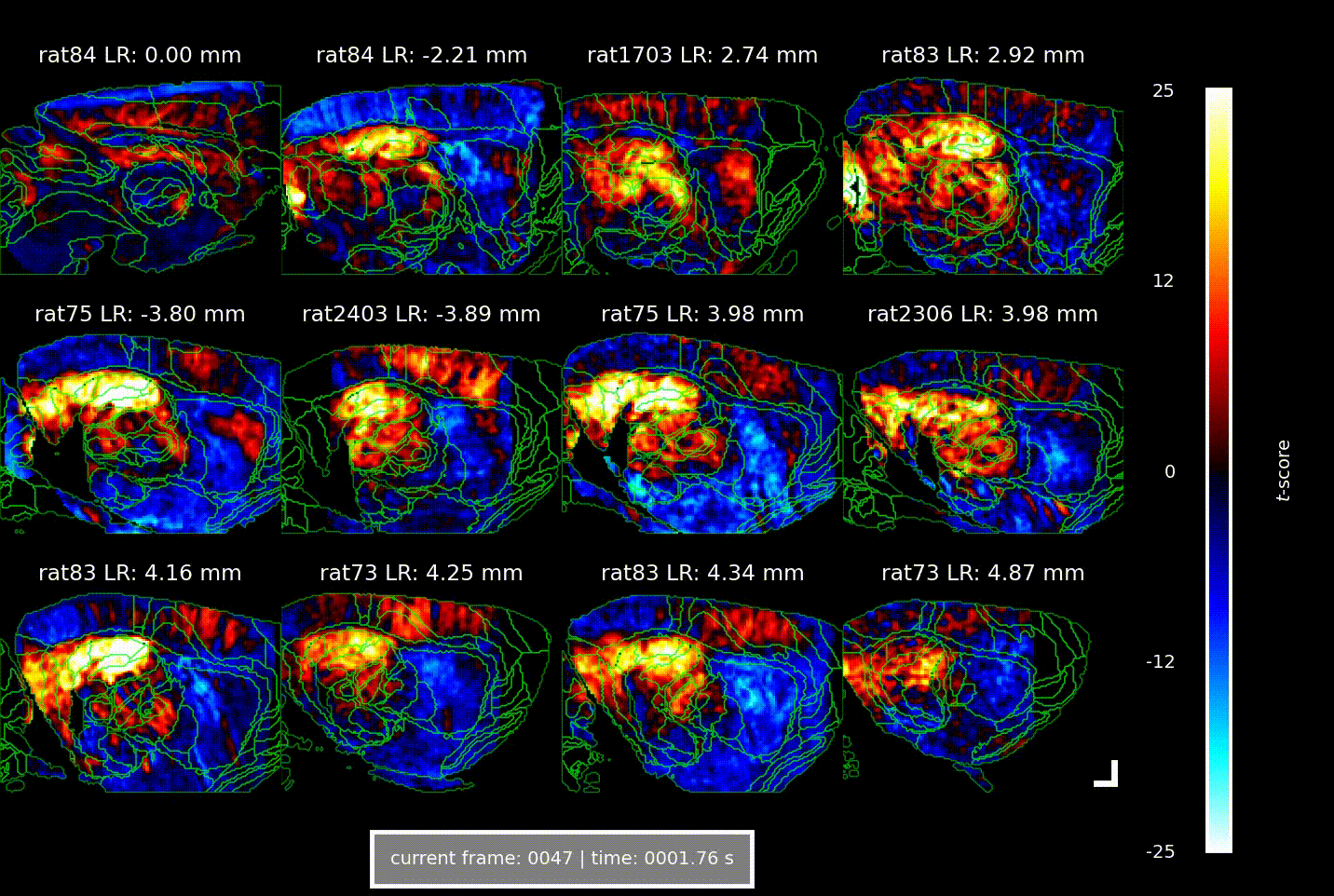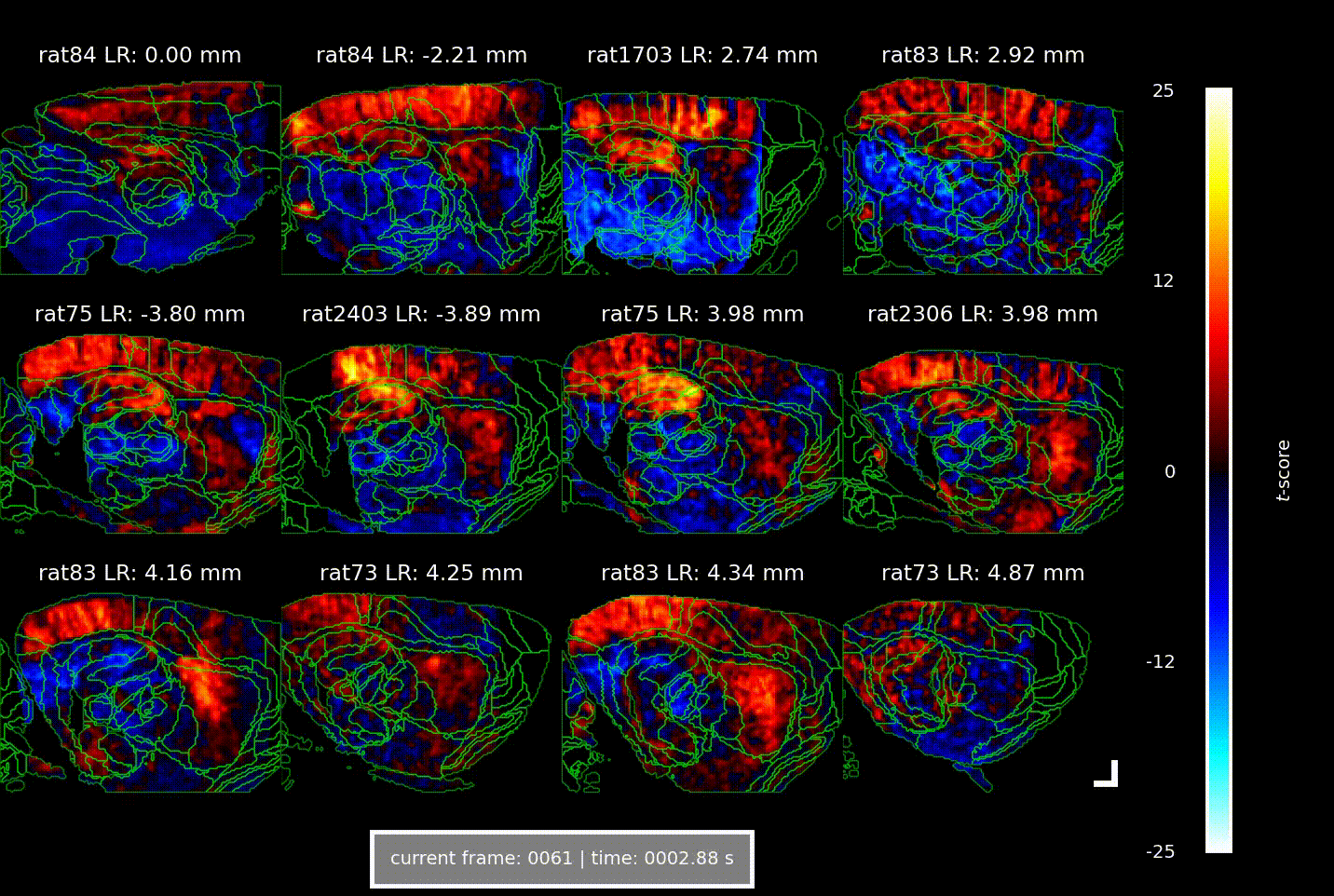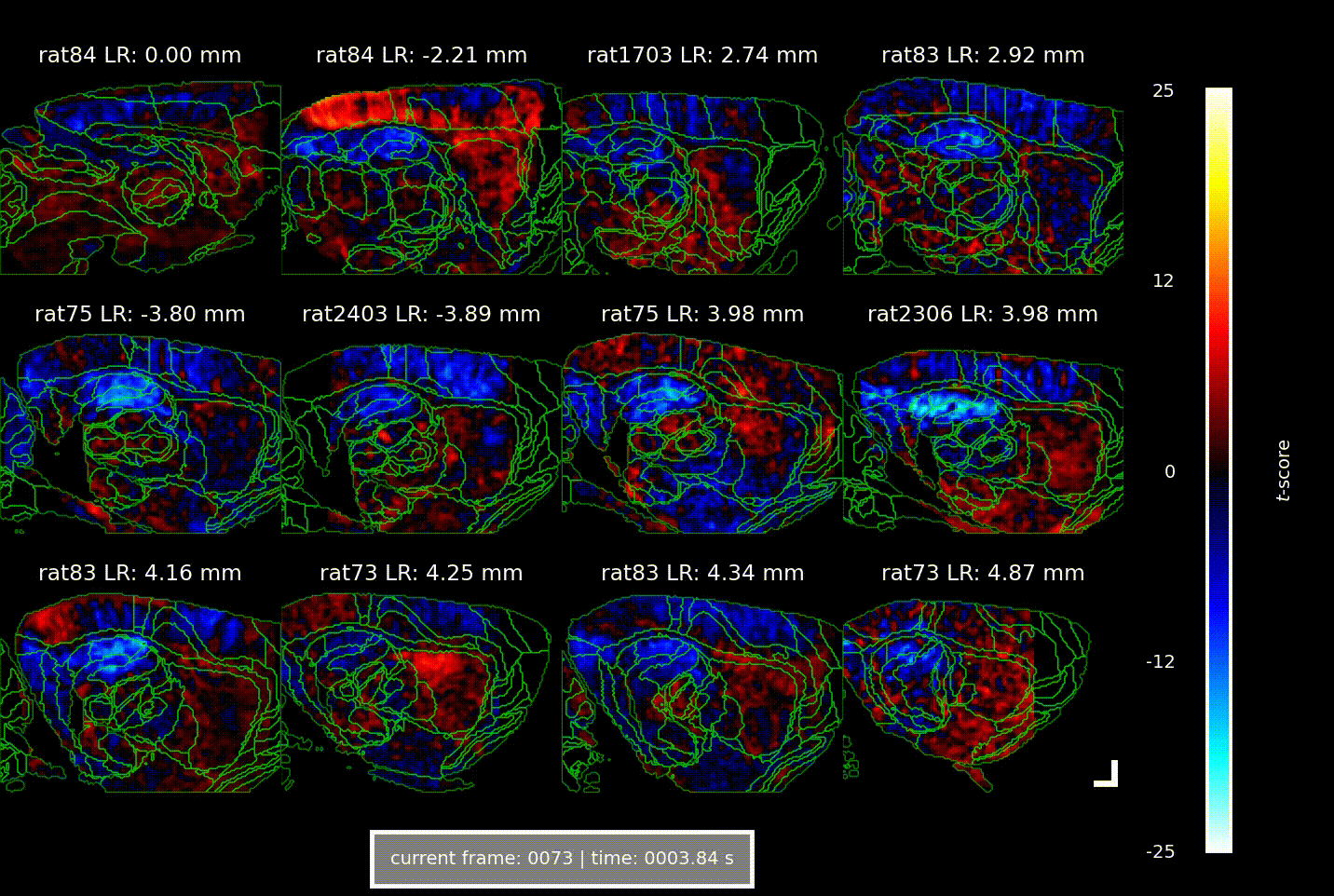
In their study, the team found that across seven animals studied in a total of about 60 imaging sessions, there were strong correlations between CBV and the animal’s speed of locomotion, as well as with its angular head velocity. Interestingly, these correlations were constrained to well-defined ‘speed-responsive’ regions, suggesting that metabolic demands are confined to specific circuits, and don’t arise from broader, non-specific activations.
These correlations also worked well in reverse, enabling animal locomotion speed (and also proximity to the arena walls) to be ‘decoded’ from the CBV response. This held true, albeit with weaker performance, even when the decoder was trained on one animal and then applied to other animals. This shows that CBV-based speed encoding is a general feature across animals, and works even when the imaging planes differ and precise anatomical registration is not possible.
Overall, the increases in CBV over time and space manifested as a ‘wave of activation’ propagating from the medial entorhinal cortex to the parahippocampus to the hippocampus, lagging behind the actual speed signal by 1.5–2.5 s – a phenomenon that the authors suggest indicates the flow of spatial information through the brain.
Videos showing the brain-wide vascular ‘signature’ of locomotion speed, acquired using Iconeus One. Each of the 12 panels shows a different combination of animal and parasagittal plane (details indicated above each panel); each frame corresponds to a different time delay (or lag) between the animal’s speed of movement and the change in CBV (indicated below); the colors indicate the strength of the statistical association (t-score, indicated at right). The analysis reveals well-defined activation patterns in the hippocampus and surrounding parahippocampal regions, with different subregions responding at distinct delays.
Remarkable insights into the neurovascular ‘signature’
The team say that because the CBV response increases in specific regions as the animal moves faster, it suggests that the energetic cost of processing spatial information also increases. The study has also provided what they describe as “the first systems-level view of how speed-related information propagates hierarchically through the complete hippocampal formation”.
Crucially, these new insights have been enabled by the capabilities of fUS. Dr Jeremy Ferrier, Head of Product Development at Iconeus and one of the report’s authors, says “To date, no other technique really combines the depth, spatial coverage, temporal resolution and behavioral freedom of fUS – and that’s why this study has been the first to reveal this remarkable neurovascular ‘signature’ of movement speed”.
The authors conclude with ideas for future experiments, including combining fUS with other techniques to understand the underlying cellular activity, and exploring whether the CBV dynamics seen in this study reflect other cognitive processes taking place during navigation, such as planning, memory, and decision-making.
Reference:
F. Cybis Pereira, S.H. Castedo, S. Le Meur-Diebolt, N. Ialy-Radio, S. Bhattacharya, J. Ferrier, B.F. Osmanski, S. Cocco, R. Monasson, S. Pezet and M. Tanter, A vascular code for speed in the spatial navigation system, Cell Reports, 2025, 45: 116791, DOI: 10.1016/j.celrep.2025.116791.