Resting-state functional connectivity
In an analogous fashion to fMRI, functional ultrasound can be used to map intrinsic brain connectivity (often called resting-state functional connectivity or resting-state networks), by detecting correlated fluctuations of spontaneous blood flow.
The high spatial and temporal resolution of fUS, and its much greater inherent sensitivity than BOLD-based fMRI, makes it a powerful way of studying neuropsychiatric diseases, and potentially aiding early diagnosis. Below are a few examples of fUS in action.
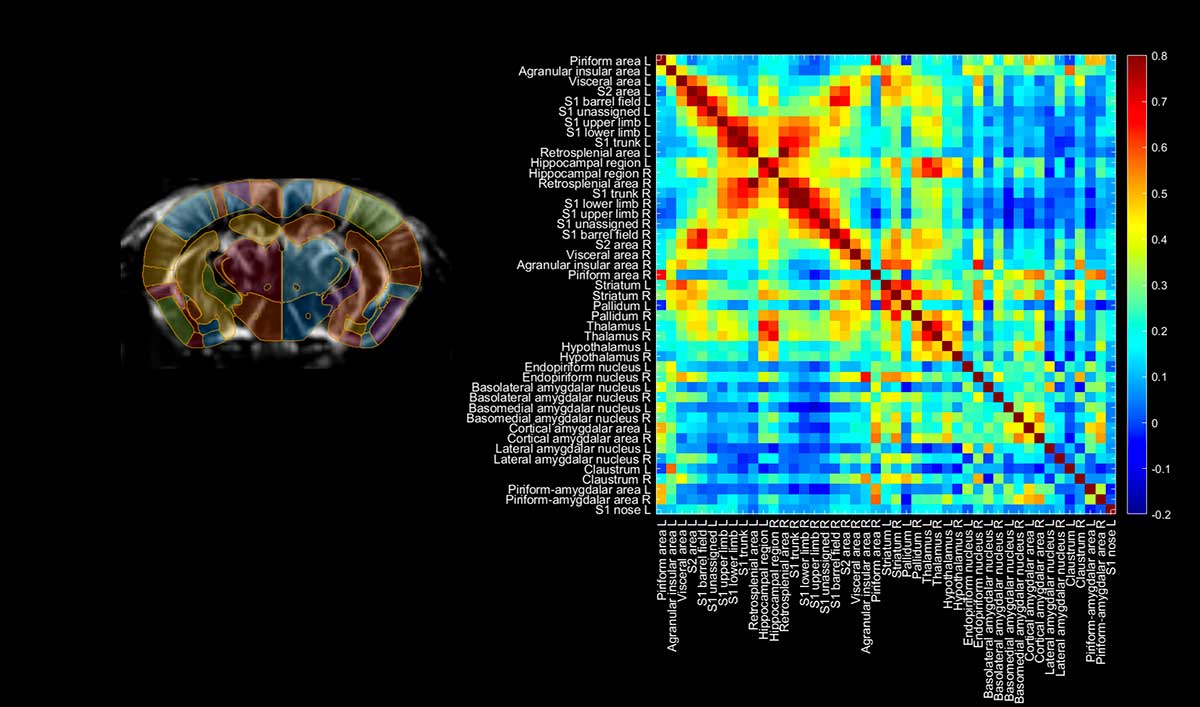
2D connectivity matrices
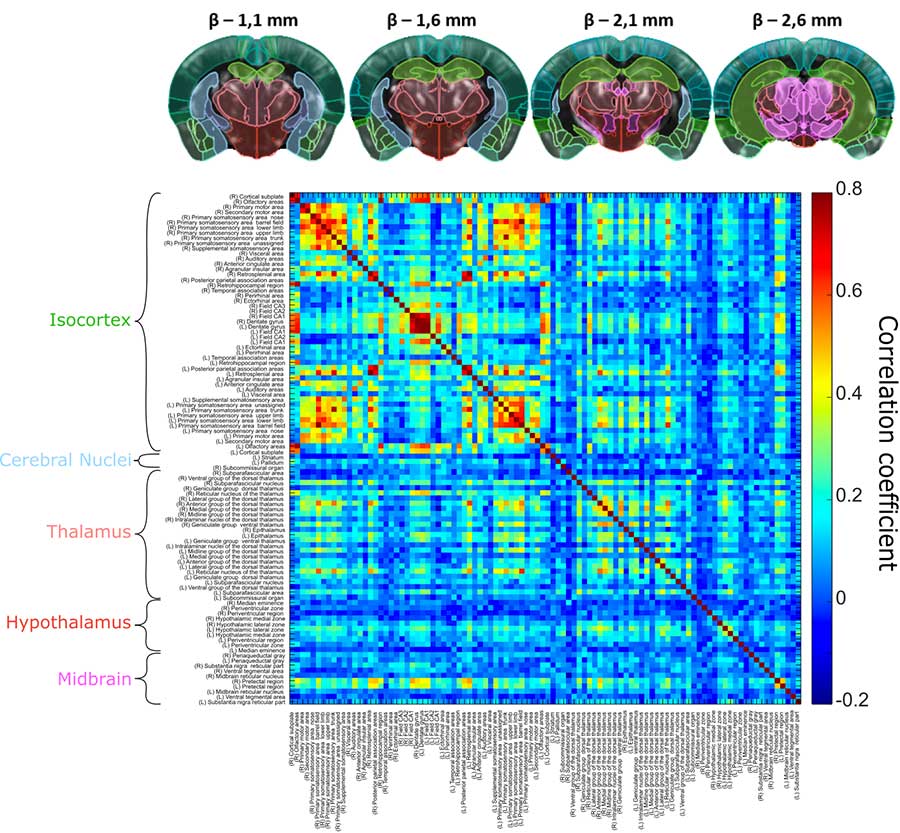
This 3D connectivity matrix for a mouse model was acquired non-invasively in 20 minutes, and shows strong interhemispheric connectivity patterns between four slices, with correlation coefficients up to 0.8. Reproduced from Bertolo et al., Journal of Visualized Experiments, 2021 (licensed under CC BY-NC-ND 3.0)
3D connectivity matrices
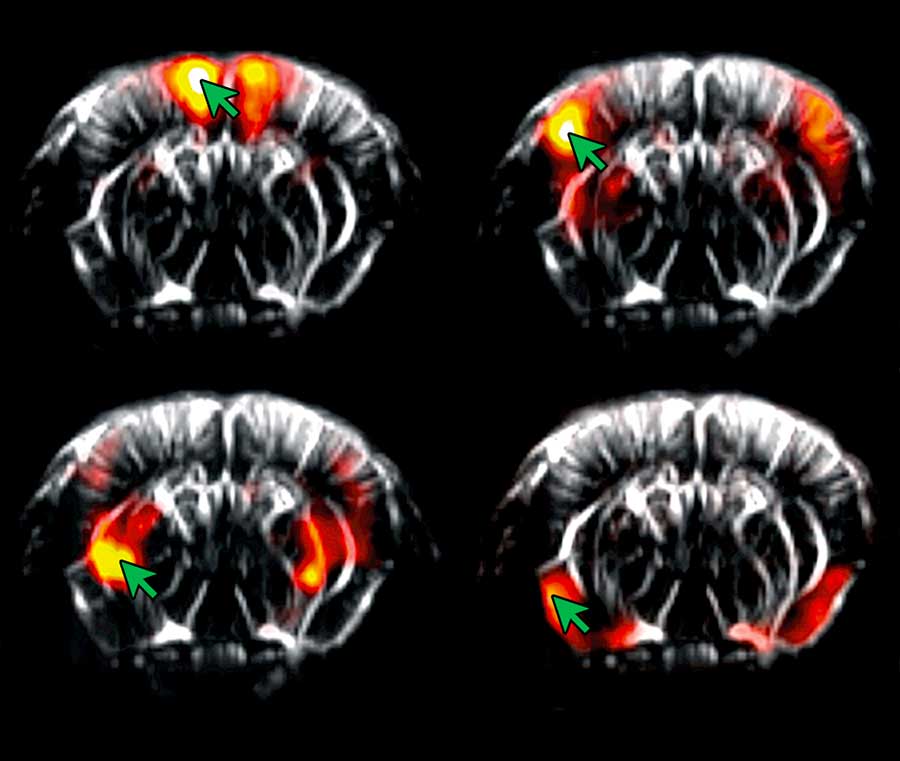
Seed-based correlation mapping
Another way of visualizing brain activity relationships is by correlating voxel activity across the whole brain with the activity of a small ‘seed’ region – known as seed-based (or ROI-based) functional connectivity.
This approach is not so dependent on the segmentation of the brain into functional regions, since you only need to define a single region.
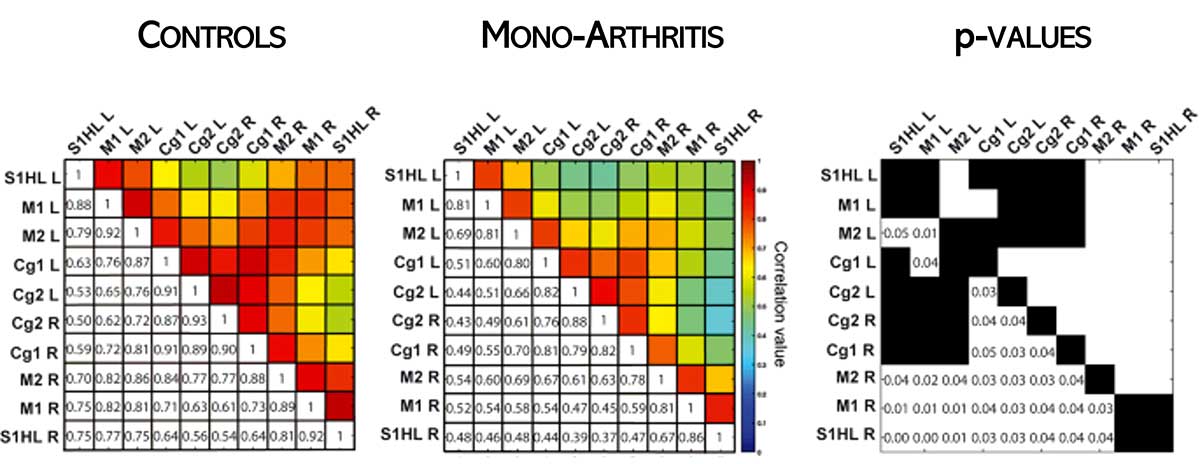
Functional connectivity in pathological models
A different manifestation of functional connectivity is looking at how differences relevant to animal pathologies influence connectivity across the whole brain.
For example, functional ultrasound has been used to study the role of oxytocin in rat pups (Mairesse et al., Glia, 2019) and to investigate sensitivity to inflammatory pain in anesthetized rats, as shown here.
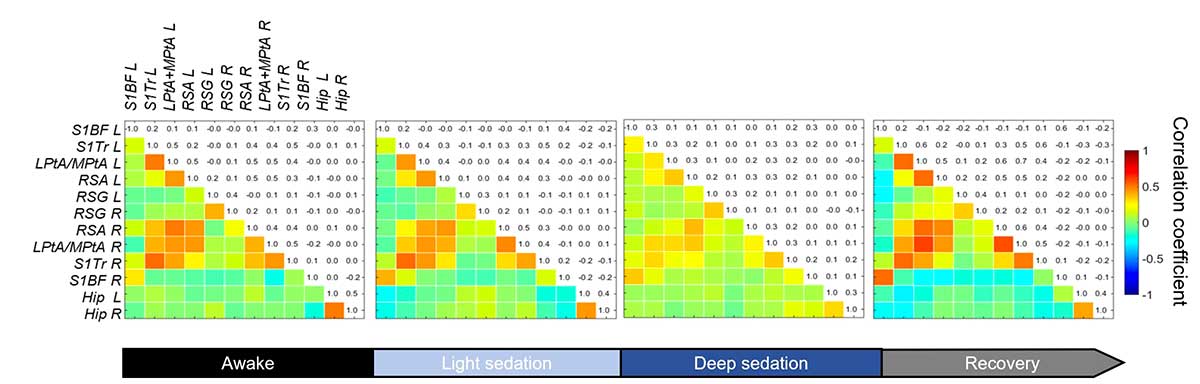
Investigating connectivity in moving animals
Connectivity matrices are normally developed for anesthetized animals, but an important development is using the light, robust Iconeus One probes to obtain results from awake mice (freely-moving or head-fixed), eliminating the bias of anesthetics.
As shown in the example below, this capability allows the effects on brain activity of anesthetics themselves to be studied, and it may also be relevant for analyzing genetically modified mice models.